Introduction
Breast cancer is a horrible disease that affects millions worldwide. In the US and other high-income countries, advances in medicine and increased awareness have significantly improved the survival rate of breast cancer to 80% or higher. However, in many lower-income countries the survival rate is below 40%, largely due to a lack of early detection systems.1
Advances in AI and medicine can make massive differences in beating diseases like breast cancer by extending diagnostics, enhancing pattern recognition in imaging, and deploying these resources for those who need them most.
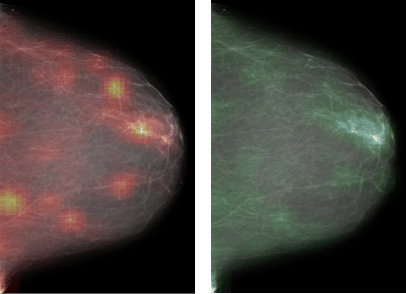
One promising advancement for early detection breast cancer systems is the application of computer vision to medical imagery. In recent years, deep learning has improved the quality of computer vision technology tremendously by learning from data. These techniques can also be applied to radiology scans to recognize and highlight possible malignant or benign areas.
At the Center of Data Science and Department of Radiology at NYU, researchers recently showed that a well trained Convolutional Neural Network (CNN), in tandem with radiologists’ predictions, delivered more accurate predictions than either by themselves.
These cutting-edge techniques and models are constantly being improved and shared, but after these models are created and trained, they need to be scaled.
Hospitals and radiology labs need to be able to productionize, scale, and deliver the results from these models so they can actually be leveraged in real-world scenarios. That’s where AI Infrastructure platforms like Pachyderm come into the picture.
Pachyderm gives you a powerful data-oriented machine learning platform that helps take research projects from the lab to enterprise-grade applications. Its immutable, version-controlled file system and Docker-based pipelines make it easy to build a robust platform capable of reaching millions across the globe.
We’ve built upon the NYU researchers’ work to show you how to scale their breast cancer detection system using three key principle techniques:
- Data Parallelism – Distribute the data so that each exam gets processed in parallel to meet any demand.
- Resource Utilization – Separate the GPU processes from CPU-only ones to maximize resource utilization and reduce operational costs.
- Collaboration – allow teams of researchers to collaborate, extend, and build upon the NYU team’s base workflow more easily.
The full source code can be found here.
Data Parallelism
Data parallelism is one of the simplest and most common ways to scale a data operation. Instead of looping through each piece of data in a single process, you run multiple instances of your code on multiple workers, and each worker can process a slice of data at the same time.
There are many ways to implement multi-threaded code to perform data parallelism, but with Pachyderm’s distributed processing model, we can autoscale our diagnostics processes dynamically as new scans arrive simply by splitting our data appropriately. Since each processing step is run in a Docker image, we can seamlessly scale out as many instances as we need and Pachyderm will automatically distribute data across all available workers.
Defining the Input Data
Pachyderm’s file system (PFS) versions data written to these repositories and sends it to the processing workers that request it. The input to our pipeline(s) are radiology exams, so we create a Pachyderm data repository (similar to a Git code repository) where these scans will be uploaded.
The CNN model relies on 4 radiological images from a single exam to predict the presence of cancer. We create a directory for each exam, identified with its (unique_exam_id). Each pipeline worker classifies a single breast exam, so we can increase the throughput of our system by adding more workers.
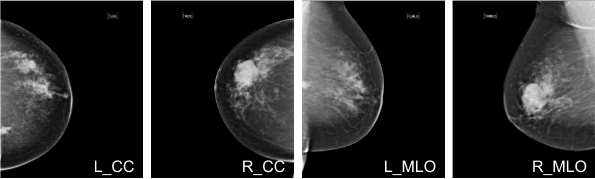
breast_exams/
├── <unique_exam_id>
│ ├── L_CC.png
│ ├── L_MLO.png
│ ├── R_CC.png
│ ├── R_MLO.png
│ └── gen_exam_list.pkl
├── <unique_exam_id>
│ ├── L_CC.png
│ ├── L_MLO.png
│ ├── R_CC.png
│ ├── R_MLO.png
│ └── gen_exam_list.pkl
...
Organizing our files in a Pachyderm data repository is similar to a file system. The pipeline can be told what directory (or files) need to be available for each worker. Note: The pkl file is metadata about the exam image list.
Every run by one of our workers uses the pre-trained model to process a single batch of images (e.g. Unique_exam_ID 1). If we have thousands of scans that need processing, they’ll be distributed automatically across all available workers.
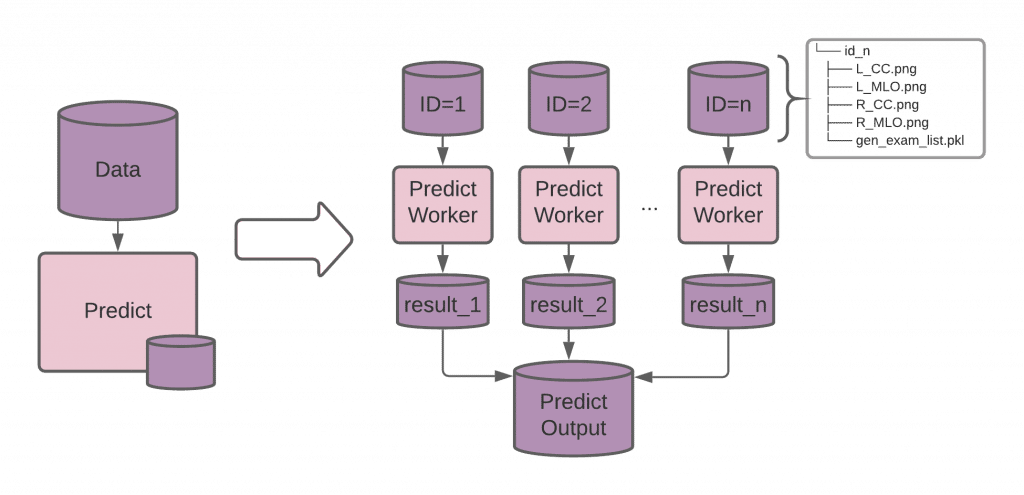
The models are placed in another data repository, models, so they can be updated independently from the Docker image. Pachyderm can detect when a new model is committed to our data repository and automatically reprocess all breast exam data with the new model(s) and update the results or know to only use the new model for incoming scans going forward.
Resource Utilization
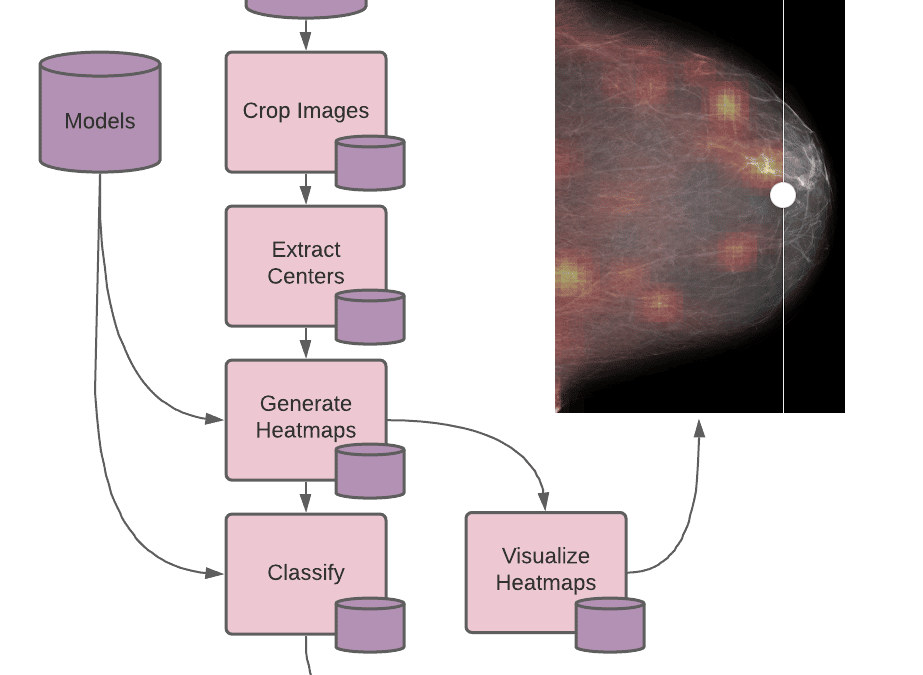
The second principle that should be considered when scaling research is resource utilization. Many deep learning techniques run most efficiently on GPUs; however, horizontally scaling GPU resources for data parallelism can be considerably expensive and even cost-prohibitive in some cases. By splitting out processes that require GPUs from the CPU-only processes, we can scale each independently, only using the GPUs when necessary. In our example, we break the NYU research team’s run.sh script into 4 discrete stages with one additional stage for our heatmap visualization (5 stages in total), and make a pipeline for each.
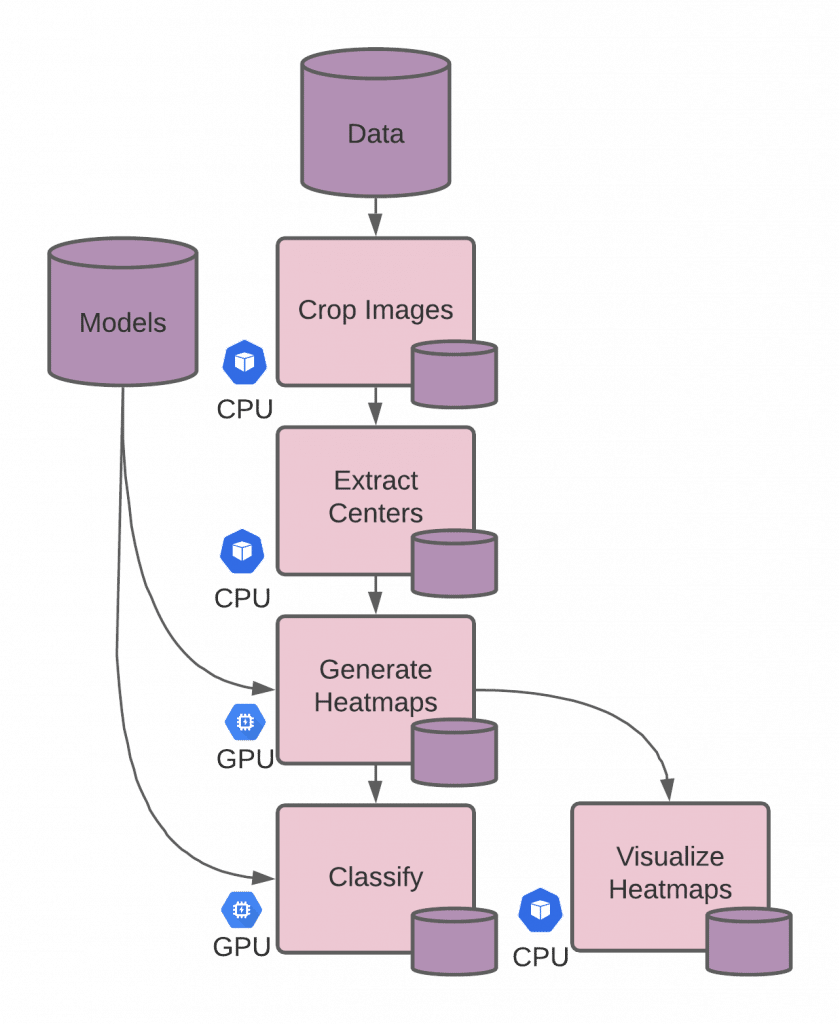
Each of these pipelines has an entrypoint script defining what code runs, the inputs required, and any additional configuration needed for the workers (such as a GPU requirement). When the pipeline executes, a worker runs the entrypoint on its data and writes output files to its own data repository (with the same name as the pipeline), which can then be read by other pipelines. The source files for each of these stages can be found in the example.
Each of these pipelines can then be scaled independently with their different resource requirements. Pachyderm handles all the dependencies and coordinates when to run jobs according to the availability of the pipeline’s required inputs.
Collaboration and Iteration
Creating a highly scalable inference framework for an enterprise healthcare app requires more than just parallelizing tasks. Oftentimes you need to parallelize your team and their time as well.
In the previous section, we split our inference code into discrete steps for resource utilization. But dividing our code into stages also allows different teams or team members to focus on a specific part of the pipeline. For example, if the “extract centers” stage is updated, perhaps to focus on a more specific area in the scan, someone can modify and update that pipeline independently from the rest of the pipelines in the project. Pachyderm monitors when pipelines are updated and optionally reprocesses all data affected with the updated code while leaving everything else untouched. It also kicks off any downstream pipeline whose input has changed, still skipping any that have not.
Pachyderm relies on changes in data as the means of communication between pipelines. Each commit in the data repository tracks which files are new or changed. This has two benefits:
- Reverting to a previous version is always possible, and
- If some datums remain unchanged pipelines are able to skip redundant computation.
In our previous example, of updating the “extract centers” pipeline, the “crop” stage would not re-run since it is upstream, but all downstream pipelines will rerun if the extracted center is different from the previous version. We use this technique to track changes in our visualization heatmaps as models change.
These features provide strong collaboration points for groups of researchers that may be working together, but on separate portions of the same project.
Conclusion
In the case of breast cancer, the quality of diagnostic predictions is essential. False positives and false negatives can have devastating real-world consequences when it comes to healthcare. A false positive in an exam leads to unnecessary and costly medical procedures and puts tremendous psychological strain on the person affected by it. Even worse, a false negative means someone doesn’t get the needed treatment which can have fatal consequences.
Advances in computer vision and AI are showing tremendous promise as we’ve seen in the case of breast cancer detection. Scaling this research is essential to delivering it to those who need it most, and with tools like Pachyderm, you can introduce parallelism techniques, decrease computation costs, and efficiently collaborate with minimal modifications to the codebase. The flexibility of the Pachyderm’ platform makes it simple to scale research effectively, able to put cutting edge diagnostics into practitioners’ hands faster.
Check out the full breast cancer detection example and other similar projects in our Bio-tech kit, or connect with us on slack to learn how to apply these productionization techniques to your life science-related use case.
This blog has been republished by AIIA. To view the original article, please click HERE.
Recent Comments